Regulation of iron uptake by hemerythrin ubiquitin ligases
The iron sensing-signalling mechanism in plants is relatively poorly understood, in contrast to the well-studied systems in bacteria, yeast and mammals. However, much progress has been made recently, highlighting the uniqueness of the putative iron-sensing proteins in plants and how they regulate the iron deficiency response.
When iron sensing is broken, we expect to see a drammatic increase in the iron concentration in a cell or in all parts of a multi-cellular organism, together with deregulated (maximally increased) expression of iron uptake genes, such as seen in the yeast aft1 mutant. Not many mutants with such a phenotype have been found in plants, except for brutus-3 (bts-3) in Arabidopsis (Hindt et al. 2017, doi: 10.1039/c7mt00152e) and dgl/bts1 in pea (Harrington et al 2024). Combined with reverse genetics studies of genes with distant homology to the mammalian FBXL5 iron/oxygen senor (Kobayashi et al. 2013, doi: 10.1038/ncomms3792; Selote et al. 2015, doi: 10.1104/pp.114.250837), the plant BRUTUS/HRZ proteins are clearly playing a key role in the iron-signalling mechanism.
The BRUTUS/HRZ proteins form a unique subfamily of E3 ligases, having a N-terminal domain with three tightly packed hemerythrin (Hr) sub-domains. This structure is not found anywhere else in biology, to our knowledge, but combined with a Pirh2/RCHY-type E3 ligase the proteins are highly conserved from green algae to higher plants (Pullin et al 2025).
Because of the novelty of the ‘triple Hr’ structure, we conducted in-depth biochemical studies to demonstrate (i) binding of 2 respectively 3 diiron centres with Hr-like properties to the N-terminal domain of BTS and BTS-Like proteins; and (ii) that iron binding is dynamic with fast kinetics, as expected for a sensor. We also showed that the dgl variant BTS1 protein is defective in iron sensing because it lacks one of the 3 diiron centres and has impaired re-oxidation activity (Pullin et al 2025). A short presentation of this paper, made more accessible for non-biochemists, can be found here.
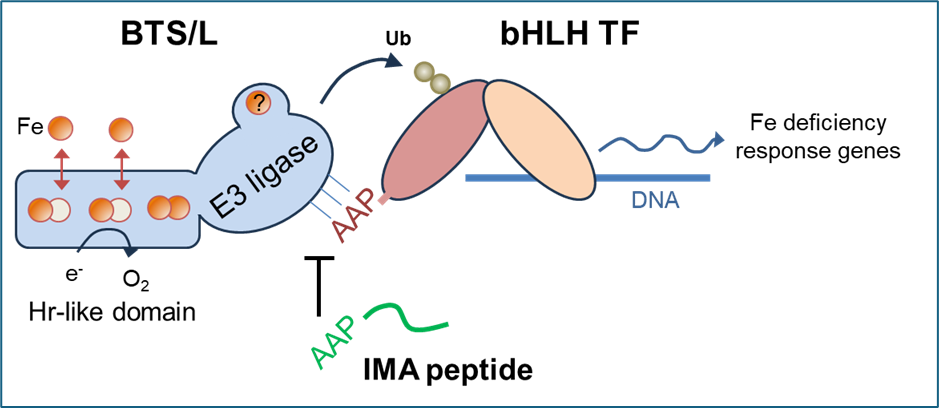
The next question is how iron binding and redox changes in the N-terminal domain of the protein modulate the E3 ligase activity of the C-terminal domain. Possibly, large conformational changes could affect interactions with the substrates, which are the E2 ubiquitin donor and the transcription factors.
The function of BTSL proteins in Arabidopsis
Phylogenetic analysis showed two separate clades in higher plants: the BRUTUS (BTS) and BRUTUS-Like (BTSL) proteins. The rice HRZ proteins belong to the BTS clade. Mutant studies have shown that BTS and HRZ are essential genes, but the BTSL genes in Arabidopsis are not.
Our in-depth functional analysis of the two redundant BTSL genes in Arabidopsis showed that the transcription factor FIT is a likely substate protein. FIT is a central player in the transcription cascade enabling the response to iron defiency, by inducing the expression of genes for iron uptake. We found that BTSL2 protein interacts directly with FIT in vivo and in vitro, and btsl1 btsl2 double mutants have sustained levels of FIT when moved from iron deficiency to resupply (Rodriguez-Celma et al 2019 PNAS).
